Lisbon-K Chromosome Dataset: Difference between revisions
No edit summary |
No edit summary |
||
| (45 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
<font size="4">Introduction</font> | |||
A new, Lisbon-K Chromosome Dataset, based on '''bone marrow''' cell chromosomes, extracted from | |||
A new, Lisbon- | |||
patients suffering from '''leukemia''', ordered and annotated by the technicians of the Institute of | patients suffering from '''leukemia''', ordered and annotated by the technicians of the Institute of | ||
Molecular Medicine of Lisbon is presented here. This data set of 200 normal ''karyograms'' with 9200 chromosomes is a very important tool from a research point of view because at last a ground truth is available to test classification and pairing algorithms for this type of cells. | Molecular Medicine of Lisbon is presented here. This data set of 200 normal ''karyograms'' with 9200 chromosomes is a very important tool from a research point of view because at last a ground truth is available to test classification and pairing algorithms for this type of cells. | ||
| Line 11: | Line 9: | ||
<div style="float:right;"> | <div style="float:right;"> | ||
[[Image:CariogramaImagemApresentacaoWikiChromosomeDataser.JPG| | [[Image:CariogramaImagemApresentacaoWikiChromosomeDataser.JPG|200px]] | ||
</div> | </div> | ||
| Line 17: | Line 15: | ||
== Background & Framework == | == Background & Framework == | ||
''Some extracts from our paper [1], regarding chromosome data (without any references)'': | ''Some extracts from our paper [1], regarding chromosome data (without any references)'': | ||
| Line 66: | Line 65: | ||
* References: | * References: | ||
** [ | ** [1] Rodrigo Ventura, Artem Khmelinskii and João Sanches, [http://dx.doi.org/10.1109/IEMBS.2010.5626237 Classifier-assisted metric for chromosome pairing], ''Proceedings of the 32nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, August 31-September 4, Buenos Aires, Argentina, 2010 Pages: 6729-6732'' | ||
** [ | ** '''[2]''' Artem Khmelinskii, Rodrigo Ventura and João Sanches, [http://dx.doi.org/10.1109/TBME.2010.2040279 '''A novel metric for bone marrow cells chromosome pairing'''], ''IEEE Transactions on Biomedical Engineering, Volume 57, Issue 6, Pages: 1420-1429, June 2010'' | ||
** [ | ** [3] Artem Khmelinskii, Rodrigo Ventura and João Sanches, [http://dx.doi.org/10.1109/IEMBS.2008.4649562 Automatic chromosome pairing using mutual information], ''Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, August 20-24, Vancouver, Canada, 2008, Pages: 1918-1921'' | ||
** [4] Artem Khmelinskii, Rodrigo Ventura and João Sanches, [http://dx.doi.org/10.1109/ISBI.2008.4541038 Chromosome pairing for karyotyping purposes using mutual information], ''Proceedings of the 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, May 14-17, Paris, France, 2008 Pages: 484-487'' | |||
== Lisbon-K1 Chromosome Dataset == | == Lisbon-K1 Chromosome Dataset == | ||
| Line 77: | Line 76: | ||
<div style="float:right;"> | <div style="float:right;"> | ||
<gallery caption=" Figure 3. Lisbon-K1 Chromosome Dataset Sample Images" widths="160px" heights="100px"> | <gallery caption=" Figure 3. Lisbon-K1 Chromosome Dataset Sample Images" widths="160px" heights="100px"> | ||
Image:ExemploImagemBoaLisbonK1ChromosomeDataset.jpg|(a). A " | Image:ExemploImagemBoaLisbonK1ChromosomeDataset.jpg|(a). A "High" quality karyogram | ||
Image:ExemploImagemMaLisbonK1ChromosomeDataset.jpg|(b). A " | Image:ExemploImagemMaLisbonK1ChromosomeDataset.jpg|(b). A "Low" quality karyogram | ||
</gallery> | </gallery> | ||
</div> | </div> | ||
| Line 85: | Line 84: | ||
* 200 ordered and chromosome class-numbered karyograms: | * 200 ordered and chromosome class-numbered karyograms: | ||
** 100 " | ** 100 "High/Medium" Quality | ||
*** | ***INSERT NUMBER Female | ||
*** | ***INSERT NUMBER Male | ||
** 100 " | ** 100 "Low" Quality | ||
*** | *** INSERT NUMBER Female | ||
*** | *** INSERT NUMBER Male | ||
* Origin: bone marrow cells collected from patients with Leukemia | * Origin: bone marrow cells collected from patients with Leukemia | ||
| Line 105: | Line 104: | ||
* Total number of chromosomes: (100*46)*2=9200 | * Total number of chromosomes: (100*46)*2=9200 | ||
* 768 x 512 TIFF format images | * 768 x 512 TIFF format images | ||
* | * INSERT NUMBER MB | ||
* Average Chromosome Size after segmenting the karyogram: 80 x 40 | * Average Chromosome Bounding Box Size in pixels after segmenting the karyogram: 80 x 40 | ||
'''Note''': The main difference between the "High" and the "Low" quality karyograms is related to the level of condensation of the chromosome, definition of the centromere position and band profile discretization/discrimination. As you can see in Fig.3.b), where a "Low" quality karyogram is presented, due to the high state of condensation it is very difficult to distinguish the band profile even for a trained expert. | |||
== Lisbon-K2 Chromosome Dataset (Future Work...) == | == Lisbon-K2 Chromosome Dataset (Future Work...) == | ||
In the future, another dataset will be build with more "real" and interesting data. i.e., karyograms extracted from cancerous cells of Leukemia patients, with all sort of chromosomal numerical and structural abnormalities. | In the future, another dataset will be build with more "real" and interesting data. i.e., karyograms extracted from cancerous cells of Leukemia patients, with all sort of chromosomal numerical and structural abnormalities. | ||
<!-- == Software == | |||
A simple algorithm to segment the karyogram, written in MATLAB™ will be included in the package. | A simple algorithm to segment the karyogram, written in MATLAB™ will be included in the package. | ||
* Input: Karyogram image | * Input: Karyogram image | ||
* Output: Cell array with the 46 chromosomes, rightly ordered | * Output: Cell array with the 46 chromosomes, rightly ordered | ||
--> | |||
== Dataset Request & Citing == | == Dataset Request & Citing == | ||
In order to follow-up the investigation interest in this area we ask the researchers interested in this dataset to send us an e-mail, with | |||
In order to follow-up the investigation interest in this area we ask the researchers interested in this dataset to send us an e-mail, with the name and the institute/research center you are affiliated to. A temporary download link will be send to you in the next few hours following the e-mail reception. | |||
To reference the dataset in any publication describing research performed using the dataset, or sets derived from the original dataset made available here please cite the following paper, in which the dataset was first presented and made public: | To reference the dataset in any publication describing research performed using the dataset, or sets derived from the original dataset made available here please cite the following paper, in which the dataset was first presented and made public: | ||
*Artem Khmelinskii, Rodrigo Ventura and João Sanches, [http://dx.doi.org/10.1109/TBME.2010.2040279 '''A novel metric for bone marrow cells chromosome pairing'''], ''IEEE Transactions on Biomedical Engineering, Volume 57, Issue 6, Pages: 1420-1429, June 2010'' | |||
Thank you and good work! | |||
== Other Chromosome Datasets == | |||
* [http://bioimlab.dei.unipd.it/Data%20Sets.htm '''BioImLab Chromosome datasets'''] | |||
== People == | == People == | ||
This dataset was built within a collaborative effort between the Institute for Systems and Robotics of the Instituto Superior Técnico of Lisbon and the Genomed Laboratory of Cytogenetics and Virology of the Institute of Molecular Medicine of Lisbon. Most of the credit would go to Sónia Santos for the selection of the karyograms, following the established criteria. | |||
This dataset was built within a collaborative effort between the Institute for Systems and Robotics of the Instituto Superior Técnico of Lisbon and the Genomed Laboratory of Cytogenetics and Virology of the Institute of Molecular Medicine of Lisbon. Most of the credit would go to Sónia Santos, Carla Souza and Paula Costa for the selection of the karyograms, following the established criteria. | |||
We would also like to thank Professor Maria do Carmo Fonseca (IMM) for all the needed support. | We would also like to thank Professor Maria do Carmo Fonseca (IMM) for all the needed support. | ||
<gallery widths="160px" heights="100px"> | <gallery widths="160px" heights="100px"> | ||
Image:LogotipoIST.jpg|[http://www.ist.utl.pt | Image:LogotipoIST.jpg|[http://www.ist.utl.pt IST] | ||
Image:LogotipoIsr.gif|[http:// | Image:LogotipoIsr.gif|[http://www.isr.ist.utl.pt ISR] | ||
Image:LogotipoIMM.jpg|[http://www.imm.ul.pt | Image:LogotipoIMM.jpg|[http://www.imm.fm.ul.pt IMM] | ||
Image:LogotipoGenomed.gif|[http://www.genomed.pt | Image:LogotipoGenomed.gif|[http://www.genomed.pt GenoMed] | ||
</gallery> | </gallery> | ||
== Authors == | |||
[http://scholar.google.nl/citations?user=vAh7J4AAAAAJ&hl=en Artem Khmelinskii] | |||
[http://users.isr.ist.utl.pt/~yoda/homepage/ Rodrigo Ventura] | |||
[http://users.isr.ist.utl.pt/~jmrs/ João Sanches] | |||
== Contact == | == Contact == | ||
For dataset request, questions, comments and suggestions on the data and the website, report bugs or typos, please contact: | For dataset request, questions, comments and suggestions on the data and the website, report bugs or typos, please contact: | ||
Latest revision as of 18:31, 17 May 2013
Introduction
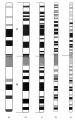
A new, Lisbon-K Chromosome Dataset, based on bone marrow cell chromosomes, extracted from patients suffering from leukemia, ordered and annotated by the technicians of the Institute of Molecular Medicine of Lisbon is presented here. This data set of 200 normal karyograms with 9200 chromosomes is a very important tool from a research point of view because at last a ground truth is available to test classification and pairing algorithms for this type of cells. The images were acquired with a Leica™ Optical Microscope DM 2500 and some image pre-processing (mainly noise reduction) and chromosome segmentation were performed with Leica™ CW 4000 Karyo software used by the clinical staff.
Background & Framework
Some extracts from our paper [1], regarding chromosome data (without any references):
"...The study of chromosomes morphology and the relation with some genetic diseases is the main goal of cytogenetics. Normal human cells have 23 classes of large linear nuclear chromosomes, in a total of 46 chromosomes per cell. This set of chromosomes contains approximately 30.000 genes (genotype) and large tracts of non coding sequences. Therefore, the examination of genetic material can involve the examination of specific chromosomal regions using DNA probes, e.g. FISH (fluorescent in situ hybridization), called molecular cytogenetics, comparative genomic hybridization (CGH) and the morphological and textural analysis of the entire chromosomes, the conventional cytogenetics, which is the focus of our work. These cytogenetics studies are very important when it comes to detection of acquired chromosomal abnormalities, such as, translocations, duplications, inversions, deletions, monosomies or trisomies that occur for example in leukemia cancerous cells and are the ideal path to take in order to characterize the different types of leukemia existent, being crucial when it comes to the right choice of treatment and follow-up for the patient, among various other applications.
The pairing of chromosomes is one of the main steps in conventional cytogenetics analysis and it is important to obtain a rightly ordered karyogram for diagnosis of genetic diseases based on the patient karyotype.
The karyogram is an image representation of stained human chromosomes with the widely used Giemsa Stain metaphase spread (G-banding) where the chromosomes are paired in 22 classes of homologous elements and two sex-determinative chromosomes (XX for the female or XY for the male), arranged in order of decreasing size. A karyotype is the set of characteristics extracted from the karyogram that may be used to detect chromosomal abnormalities. The metaphase is the step of the cellular division process where the chromosomes are at their most condensed state. In this phase the chromosomes appear well defined, allowing for the best visualization and abnormality recognition than in all the other states of the cell-division cycle.
Usually, the pairing and karyotyping procedure is done manually by visual inspection and, therefore, it is time consuming and technically demanding. After the G-banding procedure, all chromosomes gain a distinct transverse banding pattern characteristic for each class (see Figs.1, 2 and 3.a).). This banding profile is the most important feature for chromosome classification. Based on an international system for cytogenetic nomenclature (ISCN) that provides standard diagrams/ideograms of band profiles for all the chromosomes of a normal human, the clinical staff is trained to pair and interpret the karyogram according to that information. Fig.1 shows an ideogram for the chromosomes of class 1 in various states of condensation. Other features, related to the chromosome dimensions and shape are also used to increase the discriminative power of the manual or automatic classifiers.
Automatic pairing and classification is needed but it is a very difficult task. It has been an active field of research in the last two decades and still is an open problem today, namely, concerning the specific task of chromosomes pairing.
For instance, the most widely used commercial packages for cytogenetic analysis, including hardware (microscope) and software, are the Metasystems™ and Cytovision™ systems. These systems, containing state of the art algorithms for automatic detection of metaphase plates and implementation of the FISH technique, are however, still very ineffective with respect to chromosome classification and/or pairing. The same is true for the Leica™ package used by the Institute of Molecular Medicine of Lisbon (IMM) where the data used in this work was acquired..."
"...In our work a pairing algorithm for karyotyping is proposed to be used in the scope of leukemia diagnosis. For this purpose bone marrow cells are used. These chromosome images present much less quality than the ones used in the traditional genetic analysis using data sets such as Edinburgh, Copenhagen and Philadelphia, namely, concerning the centromere, band profile description/discrimination and level of chromosome condensation.
The lack of quality of the chromosome images used in the leukemia diagnostic process, when compared with other types of chromosomes images, is due to the fact that these images are based on bone marrow cells usually acquired from patients suffering from leukemia. For instance, the images from Edinburgh and Copenhagen datasets are based on routinely acquired peripheral blood cells (constitutional cytogenetics) while in the Philadelphia dataset the images are bases on cells extracted from chorionic villus (pre-natal cytogenetics). In both constitutional and pre-natal cytogenetics the observed cells are all equal, meaning that the same karyotype is always observed, independently on which cell is analyzed, making it possible to choose the metaphases that present better image quality. On the contrary, in tumoral cytogenetics (leukemia in this case), a mixture of both normal and cancerous cells is observed, with significant differences not only between normal and tumoral cells, but also within the tumoral cells, which are the key cells for the diagnosis. In addition, while in pre-natal and constitutional cytogenetics it is possible to control the cell division cycle in order to obtain chromosomes with the best morphology possible, in tumoral cytogenetics that is not possible because it is much more difficult to predict the behavior of these cancerous cells. Two different quality metaphases are displayed in Fig.2 for comparison purposes..."
- Figure 2. Metaphase Plates from Different Chromosome Datasets
-
(a). A Lisbon-K1 Chromosome Dataset Metaphase Plate
-
(b). A Copenhagen Chromosome Dataset Metaphase Plate
"...It is possible easily to observe that the chromosome images of the Lisbon-K1 Chromosome Dataset present much less quality than the ones used in the state of the art datasets described in the literature, namely with respect to the centromere, band profile discretization/discrimination and level of chromosome condensation.
The ideogram for the chromosomes of class 1 in various states of condensation in Fig.1 shows in a more comprehensive way the difference between the chromosomes used in our work and the traditional datasets. While the chromosome quality in the Edinburgh, Copenhagen and Philadelphia datasets can be included in the b). to e). interval, the quality of the chromosomes in our Lisbon-K1 Chromosome Dataset, extracted from bone marrow cells is below the a). level of band description, which can be confirmed analyzing the chromosomes of class 1 in the karyograms represented in Fig.3.
Another big difference between our dataset and the classic state of the art datasets mentioned above is the fact, that we only provide the chromosomes (displayed in the karyogram) and not the metaphases, because we are only interested in chromosomes pairing in our work, and not in chromosome segmentation..."
So, here we present a new data set, of this type of bone marrow cell chromosomes, ordered and annotated by the technicians of the Institute of Molecular Medicine of Lisbon. This data set of karyograms is a very important tool from a research point of view because at last a ground truth is available to test classification and pairing algorithms for this type of cells. The images, relevant software, and relevant information are available at this website: http://mediawiki.isr.ist.utl.pt/wiki/Lisbon-K_Chromosome_Dataset.
The images were acquired with a Leica™ Optical Microscope DM 2500 and some image pre-processing (mainly noise reduction) and chromosome segmentation were performed with Leica™ CW 4000 Karyo software used by the clinical staff. The pairing ground truth was obtained manually by the technical staff of the Institute of Molecular Medicine of Lisbon and should be used to asses the accuracy of the pairing/classification algorithms.
- References:
- [1] Rodrigo Ventura, Artem Khmelinskii and João Sanches, Classifier-assisted metric for chromosome pairing, Proceedings of the 32nd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, August 31-September 4, Buenos Aires, Argentina, 2010 Pages: 6729-6732
- [2] Artem Khmelinskii, Rodrigo Ventura and João Sanches, A novel metric for bone marrow cells chromosome pairing, IEEE Transactions on Biomedical Engineering, Volume 57, Issue 6, Pages: 1420-1429, June 2010
- [3] Artem Khmelinskii, Rodrigo Ventura and João Sanches, Automatic chromosome pairing using mutual information, Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, August 20-24, Vancouver, Canada, 2008, Pages: 1918-1921
- [4] Artem Khmelinskii, Rodrigo Ventura and João Sanches, Chromosome pairing for karyotyping purposes using mutual information, Proceedings of the 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, May 14-17, Paris, France, 2008 Pages: 484-487
Lisbon-K1 Chromosome Dataset
- Figure 3. Lisbon-K1 Chromosome Dataset Sample Images
-
(a). A "High" quality karyogram
-
(b). A "Low" quality karyogram
Description:
- 200 ordered and chromosome class-numbered karyograms:
- 100 "High/Medium" Quality
- INSERT NUMBER Female
- INSERT NUMBER Male
- 100 "Low" Quality
- INSERT NUMBER Female
- INSERT NUMBER Male
- 100 "High/Medium" Quality
- Origin: bone marrow cells collected from patients with Leukemia
- All the karyograms were selected fulfilling the following criteria:
- No structural abnormalities (such as translocations, deletions, inversions, etc.)
- No numerical abnormalities (such as monosomies or trisomies)
- No segmentation artifacts
- No artifacts related with chromosome overlapping in the metaphase plate
- All the chromosomes are correctly oriented
- Karyograms with very bended chromosomes were excluded (more than 50º)
- Without the chromosome straightening performed by the Leica™ software
- Total number of chromosomes: (100*46)*2=9200
- 768 x 512 TIFF format images
- INSERT NUMBER MB
- Average Chromosome Bounding Box Size in pixels after segmenting the karyogram: 80 x 40
Note: The main difference between the "High" and the "Low" quality karyograms is related to the level of condensation of the chromosome, definition of the centromere position and band profile discretization/discrimination. As you can see in Fig.3.b), where a "Low" quality karyogram is presented, due to the high state of condensation it is very difficult to distinguish the band profile even for a trained expert.
Lisbon-K2 Chromosome Dataset (Future Work...)
In the future, another dataset will be build with more "real" and interesting data. i.e., karyograms extracted from cancerous cells of Leukemia patients, with all sort of chromosomal numerical and structural abnormalities.
Dataset Request & Citing
In order to follow-up the investigation interest in this area we ask the researchers interested in this dataset to send us an e-mail, with the name and the institute/research center you are affiliated to. A temporary download link will be send to you in the next few hours following the e-mail reception.
To reference the dataset in any publication describing research performed using the dataset, or sets derived from the original dataset made available here please cite the following paper, in which the dataset was first presented and made public:
- Artem Khmelinskii, Rodrigo Ventura and João Sanches, A novel metric for bone marrow cells chromosome pairing, IEEE Transactions on Biomedical Engineering, Volume 57, Issue 6, Pages: 1420-1429, June 2010
Thank you and good work!
Other Chromosome Datasets
People
This dataset was built within a collaborative effort between the Institute for Systems and Robotics of the Instituto Superior Técnico of Lisbon and the Genomed Laboratory of Cytogenetics and Virology of the Institute of Molecular Medicine of Lisbon. Most of the credit would go to Sónia Santos, Carla Souza and Paula Costa for the selection of the karyograms, following the established criteria. We would also like to thank Professor Maria do Carmo Fonseca (IMM) for all the needed support.
Authors
Contact
For dataset request, questions, comments and suggestions on the data and the website, report bugs or typos, please contact:
Artem Khmelinskii
e-mail: artkhmelinskii (##) isr.ist.utl.pt